| 病毒感染严重威胁人类健康。利用人类胚胎干细胞和多能干细胞分化不同组织细胞/类器官研究病毒感染,可以克服传统细胞系对复杂的人体细胞和器官系统建模的局限。我组主要从事病毒与宿主相互作用的机制研究。利用组织细胞和类器官的干细胞分化,病理分析和药物筛选平台,研究病毒感染,探索潜在的传染病防治策略。 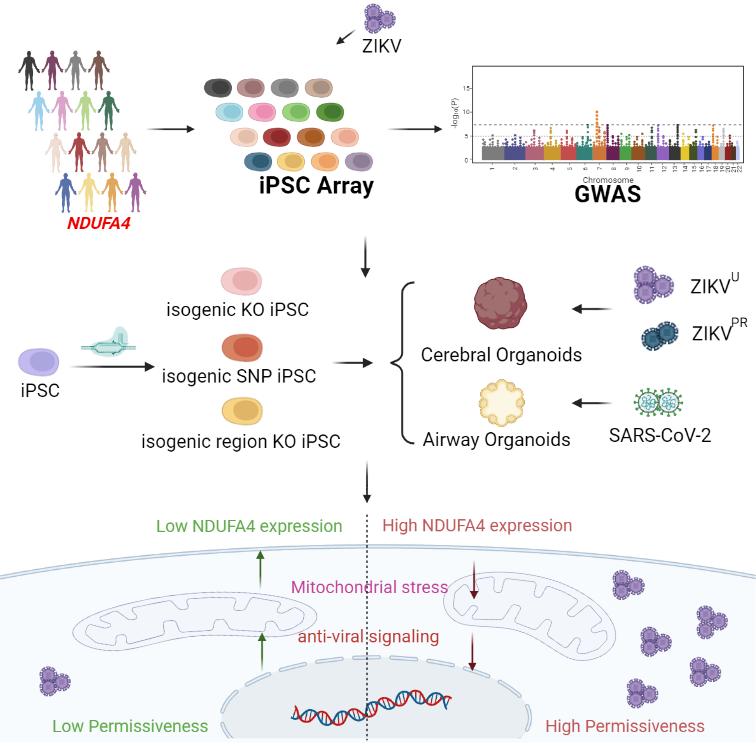
遗传变异对病毒易感性的影响
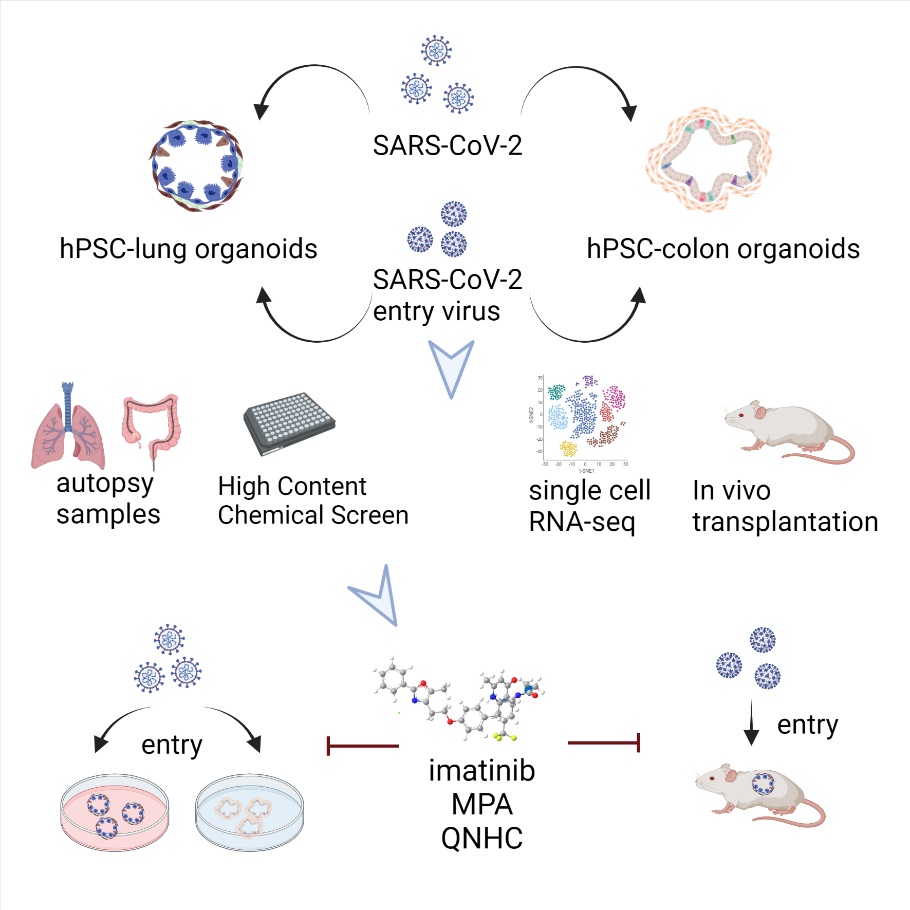
基于类器官的病毒感染和药物筛选
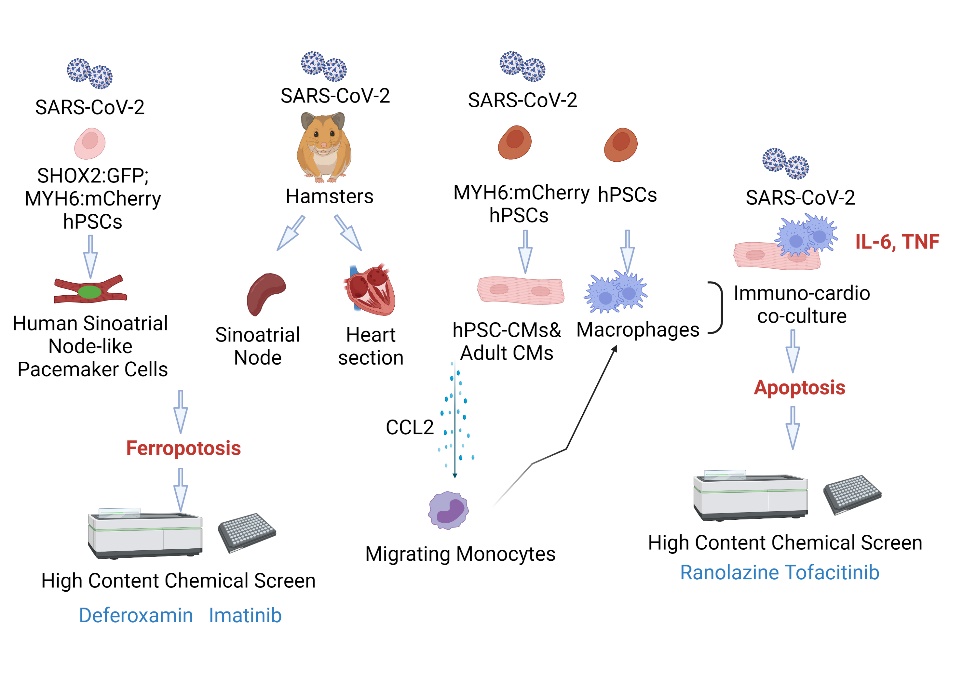
病毒感染引起宿主损伤的机制及细胞保护性药物鉴定
| 研究内容和目标:
我组将基于前期类器官分化和病毒学研究基础,深入研究:1,遗传与变异对疾病进程的影响; 2,构建复杂类器官的生理病理模型; 3.开发与鉴定潜在的传染病防治药物。 代表性发表论文: - Han Y#, Duan X#, Yang L#, Nilsson-Payant BE#, Wang P#, Duan F#, Tang X#, Yaron TM#, Zhang T, Uhl S, Bram Y, Richardson C, Zhu J, Zhao Z, Redmond D, Houghton S, Nguyen DT, Xu D, Wang X, Jessurun J, Borczuk A, Huang Y, Johnson JL, Liu Y, Xiang J, Wang H#, Cantley LC*, tenOever BR*, Ho DD*, Pan FC*, Evans T*, Chen HJ*, Schwartz RE* and Chen S*. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021; 589: 270-275
- Wang X#, Xuan Y#, Han Y#, Ding X, Ye K, Yang F, Gao P, Goff SP and Gao G*. Regulation of HIV-1 Gag-Pol Expression by Shiftless, an Inhibitor of Programmed -1 Ribosomal Frameshifting. Cell. 2019; 176:625-635 e14.
- Han Y#, Tan L#, Zhou T#, Yang L#, Carrau L#, Lacko LA, Saeed M, Zhu J, Zhao Z, Nilsson-Payant BE, Lira Neto FT, Cahir C, Giani AM, Chai JC, Li Y, Dong X, Moroziewicz D, Team NGSCA, Paull D, Zhang T, Koo S, Tan C, Danziger R, Ba Q, Feng L, Chen Z, Zhong A, Wise GJ, Xiang JZ, Wang H, Schwartz RE, tenOever BR, Noggle SA, Rice CM, Qi Q, Evans T and Chen S*. A human iPSC-array-based GWAS identifies a virus susceptibility locus in the NDUFA4 gene and functional variants. Cell Stem Cell. 2022; 29:1475-1490 e6.
- Yang L#, Han Y#, Nilsson-Payant BE#, Gupta V#, Wang P, Duan X, Tang X, Zhu J, Zhao Z, Jaffre F, Zhang T, Kim TW, Harschnitz O, Redmond D, Houghton S, Liu C, Naji A, Ciceri G, Guttikonda S, Bram Y, Nguyen DT, Cioffi M, Chandar V, Hoagland DA, Huang Y, Xiang J, Wang H, Lyden D, Borczuk A, Chen HJ, Studer L, Pan FC*, Ho DD*, tenOever BR*, Evans T*, Schwartz RE* and Chen S*. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 2020; 27:125-136 e7. Best of Cell Stem Cell 2020.
- Han Y#, Zhu J#, Yang L#, Nilsson-Payant BE#, Hurtado R, Lacko L, Sun X, Gade AR, Higgins CA, Sisso WJ, Dong X, Wang M, Zhengming Chen, Ho DD, Pitt GS, Schwartz RE*, tenOever BR*, Evans T* and Chen S*. SARS-CoV-2 infection Induces Ferroptosis of Sinoatrial Node-like Pacemaker Cells. Circ Res. 2022 130:963-977. Circulation Research Best Manuscript 2022.
- Han Y#, Yang L#, Lacko L and Chen S*. Human Organoid Models to Study SARS-CoV-2 Infection. Perspective, Nature Methods. 2022 4:418-428.
- Yang L#, Han Y#, Jaffre F#, Nilsson-Payant BE#, Bram Y#, Wang P, Zhu J, Zhang T, Redmond D, Houghton S, Uhl S, Borczuk A, Huang Y, Richardson C, Chandar V, Acklin JA, Lim JK, Chen Z, Xiang J, Ho DD, tenOever BR*, Schwartz RE*, Evans T* and Chen S*. An Immuno-Cardiac Model for Macrophage-Mediated Inflammation in COVID-19 Hearts. Circ Res. 2021 129:33-46. Cover story
- Yang L#, Kim TW#, Han Y#, Nair MS#, Harschnitz O#, Zhu J#, Wang P#, Koo SY, Lacko LA, Chandar V, Bram Y, Zhang T, Zhang W, He F, Pan C, Wu J, Huang Y, Evans T, van der Valk P, Titulaer MJ, Spoor JKH, Furler O'Brien RL, Bugiani M, W DJVdB, Schwartz RE*, Ho DD*, Studer L* and Chen S*. SARS-CoV-2 infection causes dopaminergic neuron senescence. Cell Stem Cell. 2024.
- Yang L#, Nilsson-Payant BE#, Han Y#, Jaffré F#, Zhu J#, Wang P, Zhang T, Redmond D, Houghton S and Møller R, Hoagland D, Carrau L, Horiuchi S, Goff M, Lim JK, Bram Y, Richardson C, Chandar V, Borczuk A, Huang Y, Xiang J, Ho DD*, Schwartz RE*, tenOever BR, Evans T* and Chen S*. Cardiomyocytes Recruit Monocytes upon SARS-CoV-2 Infection by Secreting CCL2. Stem Cell Reports. 2021 Jul 20.
- Yang L#, Han Y#, Zhou T#, Lacko LA, Saeed M, Tan C, Danziger R, Zhu J, Zhao Z, Cahir C, Giani AM, Li Y, Dong X, Moroziewicz D, Team NGSCA, Paull D, Chen Z, Zhong A, Noggle SA, Rice CM, Qi Q, Evans T and Chen S*. Isogenic human trophectoderm cells demonstrate the role of NDUFA4 and associated variants in ZIKV infection. iScience. 2023;26:107001.
|

