| 研究组未来的研究方向主要集中在以下三个方面: 1. 开发源头创新的新型基因工程技术。 ZFN、TALEN和CRISPR技术乃至分子生物学中广泛运用的限制性内切酶和DNA聚合酶等工具酶大部分来自于不同物种的功能基因研究。随着基因组学的发展,大量新物种的基因组完成测序和组装,大量编码新型蛋白的基因得以被发现,但其功能未知。我们希望通过生物信息学分析,找到可能成为基因工程技术新工具的候选酶,然后利用体内和体外的高通量功能检测平台筛选出具有新功能或更优特性的工具酶。通过这些研究,我们希望能够建立基因组改造的新技术,为生物学研究和疾病治疗提供更为有力的工具。 2. 在T细胞和造血干细胞中建立和优化基因编辑技术,开发新型疾病治疗方法。 嵌合抗原受体修饰(chimeric antigen receptor,CAR)T 细胞疗法作为全新的肿瘤治疗方法在临床上凸显出广阔的应用前景。这些以T细胞为基础的疗法和基因编辑相结合具有重大的应用价值。因此我们在T细胞中优化基因编辑技术,建立新型的细胞治疗方法。同时我们也在T细胞中建立进行高通量遗传学和小分子筛选的技术,以期为免疫治疗发现新的治疗靶点和药物。 3. 利用人类胚胎干细胞技术和基因编辑技术研究早期胚胎发育中的X染色体失活现象的分子机制。 X染色体失活是哺乳动物雌性在胚胎早期发育过程中对于X染色体基因表达剂量补偿的重要表观遗传调控机制,对于雌性的正常发育和生理状态起着至关重要的作用。通过优化人类naive胚胎干细胞培养和分化体系,我们建立了在体外模拟人类X染色体失活过程的研究平台。利用这个平台结合CRISPR-Cas9基因筛选技术,我们将系统研究人类X染色体失活的遗传学分子机制。 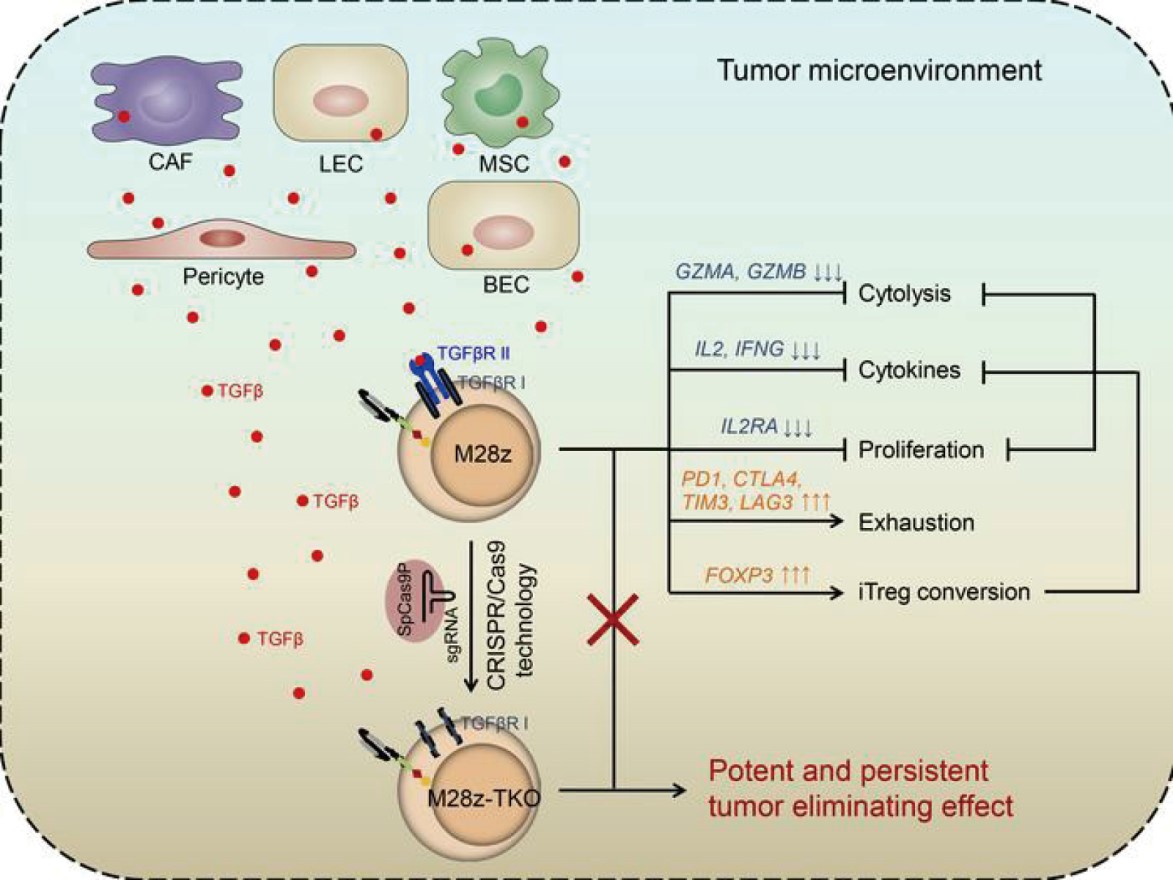
图一 利用CRISPR技术阻断TGFβ信号通路使得CAR-T清除实体瘤能力极大改善的可能机制
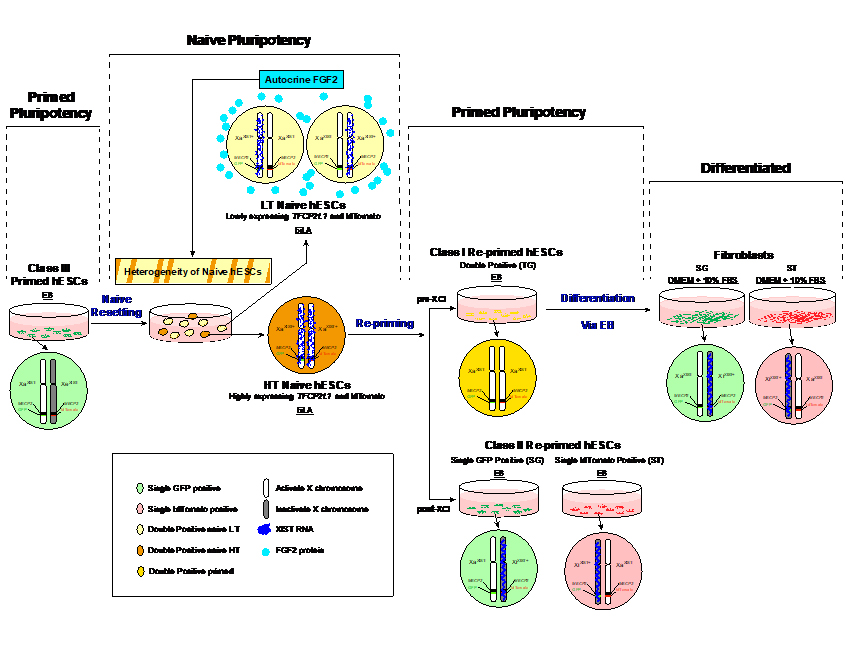
图二 模拟人类X染色体随机失活的干细胞模型
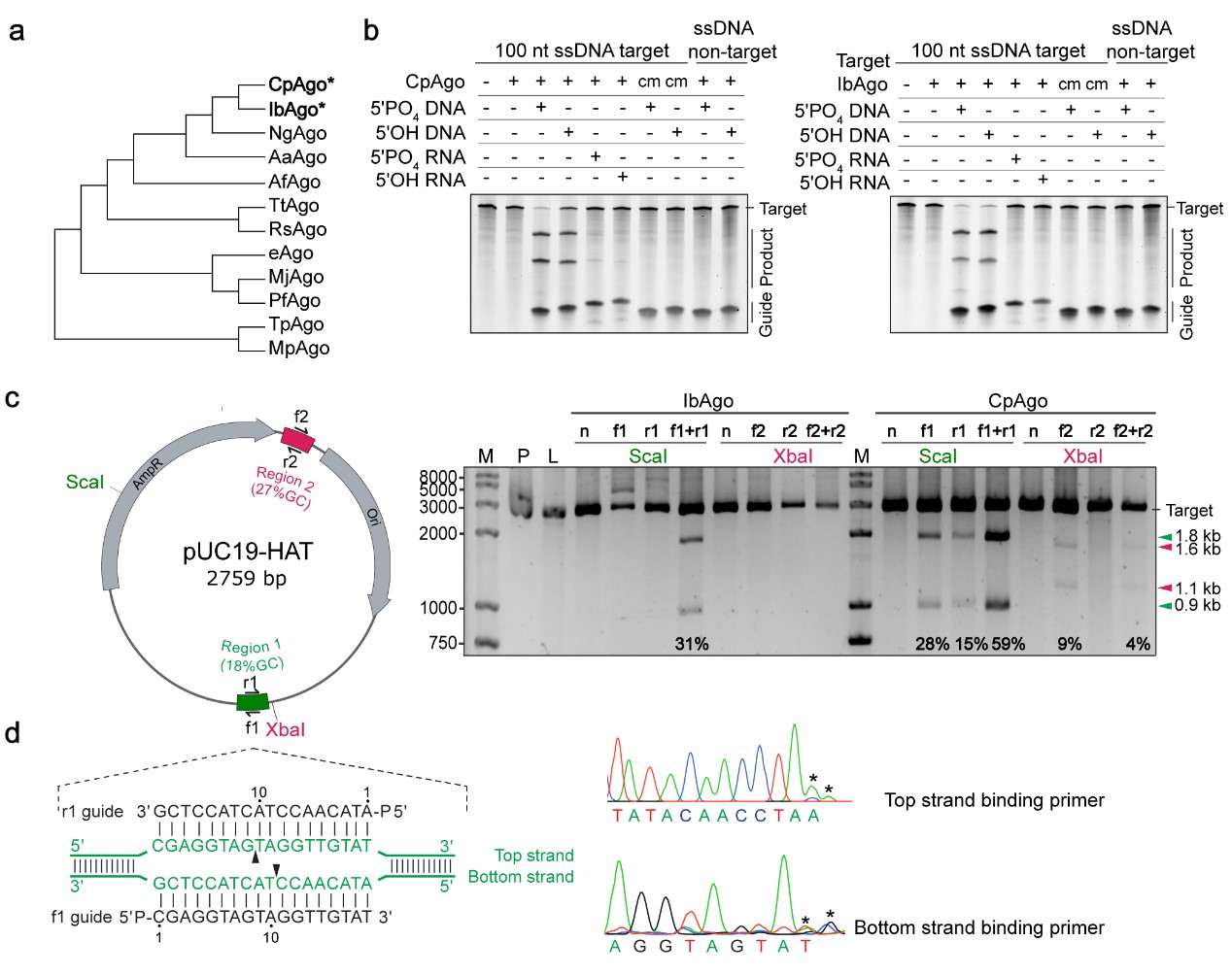
图三 CpAgo和IbAgo的DNA切割活性鉴定
| 研究内容和目标:
高效和精确的基因工程技术对于生物医学的研究和应用具有极其重要的价值。研究组的科研兴趣在于开发和应用最新的基因工程技术,在哺乳动物细胞中进行精确的基因修饰。一方面优化应用已有的基因编辑技术在T细胞和造血干细胞中进行基因改造,建立新型细胞治疗方法;另一方面挖掘开发全新的基因工程工具,建立全新基因组改造技术。同时我们也针对特定干细胞生物学基础问题进行研究。研究组研究方向主要集中在以下三个方面:
一, 开发源头创新的新型基因工程技术。
二, 在T细胞和造血干细胞中建立和优化基因编辑技术,开发新型疾病治疗方法。
三, 利用人类胚胎干细胞技术和基因编辑技术研究早期胚胎发育中的X染色体失活现象的分子机制。 代表性发表论文(*共同第一作者, #通讯作者): - An C*, Feng G*, Zhang J, Cao S, Wang Y, Wang N, Lu F, Zhou Q, Wang H#. “Overcoming Autocrine FGF Signaling-Induced Heterogeneity in Naive Human ESCs Enables Modeling of Random X Chromosome Inactivation” Cell Stem Cell, 2020 Sep;27(3):482–497.
- Tang N*, Cheng C*, Zhang X, Qiao M, Li N, Mu W, Wei X, Han W, Wang H#. “TGFβ Inhibition via CRISPR Promotes the Long-Term Efficacy of CAR-T Cells Against Solid Tumors” JCI Insight, 2020 Feb;5(4): e133977.
- Cao Y*, Sun W*, Wang J, Sheng G, Xiang G, Zhang T, Shi W, Li C, Wang Y, Zhao F#, Wang H#. “Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double- stranded DNA at 37 °C.” Cell Discovery, 2019 Jul 30;5:38.
- Wang H#,Yang H#. “Gene-edited babies: What went wrong and what could go wrong.” PLOS Biology, 2019 Apr 30;17(4):e3000224.
- Mu W*, Tang N*, Cheng C*, Sun W, Wei X, Wang H#. “In vitro transcribed sgRNA causes cell death by inducing interferon release.” Protein & Cell,2019 Jun;10(6):461-465.
- Cheng C*, Tang N*, Li J, Cao S, Zhang T, Wei X, Wang H#. “ Bacteria-free minicircle DNA system to generate integration-free CAR-T cells.” Journal of Medical Genetics, 2019 Jan;56(1):10-17.
- Xiang G*, Ren J*, Hai T*, Fu R, Yu D, Wang J, Li W, Wang H#, Zhou Q#. “Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs.” Cellular and Molecular Life Sciences. 2018 Dec;75(24):4619-4628.
- Xiang G, Wang H#. “Extended pluripotent stem cells facilitate mouse model generation.” Protein & Cell. 2018 Aug 21.doi: 10.1007/s13238-018-0573-0.
- Mu W, Zhang Y, Xue X, Liu L, Wei X, Wang H#.” 5′ capped and 3′ polyA-tailed sgRNAs enhance the efficiency of CRISPR-Cas9 system.” Protein & Cell, 2018.Jun;10(3):223–228.
- Zhang Y, Mu W, Wang H#. “Gene editing in T cell therapy.” Journal of Genetics and Genomics, 2017 Sep;44 (9) :415-422.
- Wang H#. “Editing Base in Mouse Model.” Protein & Cell, 2017 Aug;8(8):558-559.
- Wang W, Zhang Y, Wang H#. “Generating Mouse Models Using Zygote Electroporation of Nucleases (ZEN) Technology with High Efficiency and Throughput.” Methods in Molecular Biology, 2017;1605:219-230.
- Zhang Y, Zhang X, Cheng C, Mu W, Liu X, Li N, Wei X, Liu X, Xia C, Wang H#. “CRISPR-Cas9 mediated LAG-3 disruption in CAR-T cells.” Frontiers of Medicine, 2017 Dec;11(4):554-562.
- Xiang G, Zhang X, An C, Cheng C, Wang H#. “Temperature effect on CRISPR-Cas9 mediated genome editing.” Journal of Genetics and Genomics, 2017 Apr ;44(4):199-205.
- Liu X, Zhang Y, Cheng C, Cheng WA, Zhang X, Li N, Xia C, Wei X, Liu X, Wang H#. “CRISPR-Cas9 mediated multiplex gene editing in CAR-T cells.” Cell Research, 2017 Jan;27(1):154-157.
- Burgess S, Cheng L#, Gu F, Huang J, Huang Z, Lin S#, Li J, Li W, Qin W, Sun W, Songyang Z, Wei W#, Wu Q, Wang H#, Wang X, Xiong JW, Xi J, Yang H, Zhou B, Zhang B. “Questions about NgAgo” Protein Cell, 2016 Dec;7(12):913-915.
- Wang W,Kutny PM, Byers SL, Longstaff CJ, DaCosta MJ, Pang C, Zhang Y, Taft RA, Buaas FW, Wang H#. “Delivery of Cas9 protein into mouse zygotes through a series of electroporation dramatically increased the efficiency of model creation.” Journal of Genetics and Genomics, 2016 May 20;43(5):319-27.
- Qin W, Kutny PM, Maser RS, Dion SL, Lamont JD, Zhang Y, Perry GA, Wang H#.“Generating Mouse Models Using CRISPR-Cas9 Mediated Genome Editing.”Current Protocols in Mouse Biology, 2016 Mar 1;6(1):39-66.
- Cheng AW*#, Jillette N*, Lee P, Plaskon D, Fujiwara Y, Wang W, Taghbalout A, Wang H#. “Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling.” Cell Research, 2016 Feb;26(2):254-7.
- Wiles MV, Qin W, Cheng AW, Wang H#. “CRISPR-Cas9 mediated genome editing and guide RNA design.” Mammalian Genome, 2015 Oct;26(9-10):501-10.
- Qin W, Dion SL, Kutny PM, Zhang Y, Cheng AW, Jillette NL, Malhotra A, Geurts AM, Chen YG, Wang H#. “Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease.” Genetics, 2015 Jun;200(2):423-30.
- Theunissen T*, Powell B*, Wang H*, Mitalipova M, Faddah D, Reddy J, Fan Z, Maetzel D, Ganz K, Shi L, Lungjangwa T, Imsoonthornruksa S, Stelzer Y, Rangarajan S, D’Alessio A, Zhang J, Gao Q, Dawlaty M, Young R, Gray N, Jaenisch R. “Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency.”Cell Stem Cell, 2014 Oct 2; 15(4):471-87.
- Yang H, Wang H, Jaenisch R. “Generating genetically modified mice using CRISPR/Cas-mediated genome engineering.” Nature protocols, 2014 Aug; 9(8):1956-68.
- Maetzel D*,Sarkar S*, Wang H*, Mosleh LA, Cheng AW, Xu P, Gao Q, Mitalipova M, Jaenisch R. “Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells.” Stem Cell Reports, 2014 May 15; 2(6):866-80.
- Cheng AW*, Wang H*, Yang H, Shi L, Katz Y, Rangarajan S, Theunissen TW, Shivalila CS, Dadon DB, Jaenisch R. “Multiplexed activation of endogenous genes by CRISPR-on, a RNA-guided transcriptional activator system.” Cell Research, 2013 October 1; 23(10): 1163-1171.
- Yang H*, Wang H*, Shivalila CS*, Cheng AW, Shi L, Jaenisch R. “One-step generation of mice carrying reporter and conditional allele by CRISPR/Cas mediated genome editing” Cell, 2013 September 12; 154(6):1370-1379.
- Faddah D, Wang H, Buganim Y, Cheng AW, Jaenisch R. “Expression of Nanog is biallelic and equally variable as other pluripotency factors.” Cell Stem Cell, 2013 Jul 3;13(1):23-9.
- Wang H*, Hu YC*, Markoulaki S, Welstead GG, Shivalila CS, Cheng AW, Pyntikova T, Dadon D, Voytas DF, Bogdanove AJ, Page DC, Jaenisch R. “TALEN-mediated editing of the Mouse Y Chromosome.” Nature Biotechnology, 2013 Jun; 31(6):530-2.
- Wang H*, Yang H*, Shivalila CS*, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. “One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas mediated genome engineering.” Cell, 2013 May 9;153(4):910-8.
|

