| 猪是重要的经济动物,在世界范围内为人类提供了最主要的肉食来源。但由于良种猪资源大量需要进口以及育种技术的落后,使得我国养猪产业链极为脆弱,严重制约养猪产业的健康和可持续发展。同时猪也是构建人类疾病动物模型和研究再生医学的合适动物,在生物医学中有广泛的应用。通过基因组编辑(Genome Editing, GE)技术针对影响猪经济性状的功能基因进行精确和多样化的修饰为猪的品种培育、性能改良和功能基因挖掘提供了有效的解决方案。但由于猪基因学研究的相对滞后和没有功能明确的干细胞可以利用,猪的基因组编辑一直存在效率低,周期长等困难。针对上述科学问题,实验室采用正向遗传学和反向遗传学的研究策略,主要研究方向为:1. 通过化学诱变进行猪全基因组水平的随机突变和表型分析,建立突变家系,筛选功能基因和培育育种新材料;2. 通过GE方法进行猪的基因组遗传修饰进而改善猪的农业生产性状或构建生物医学研究的大动物模型;3,猪早期胚胎发育的表观遗传调控和体细胞核移植。目前的研究成果主要包括通过化学诱变建立了较大规模的突变体,筛选到了影响骨骼发育、肌肉发生、血细胞再生、行为、毛色等猪重要性状的功能基因和突变家系;建立了高效的猪GE平台,获得了携带有人抗凝血因子9, Furin蛋白和α-抗胰蛋白酶的转基因猪,单基因敲除,多基因同时敲除,定点插入,定点突变等多种类型基因修饰猪,不但为进一步培育猪新品种提供了育种材料和分子标记,也促进了猪在生物医学中的应用。目前承担的科研项目主要有中国科学院知识创新工程前沿领域项目、国家重点研发计划、国家杰出青年科学基金、国家自然基金等。 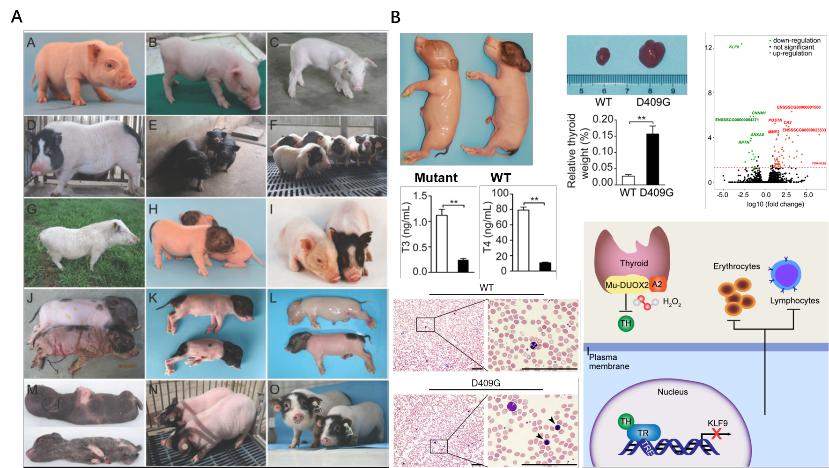
利用ENU化学诱变技术高效地创制了大量大动物模型,并且成功解析了一例模拟人类先天性甲状腺功能低下的大动物疾病模型。
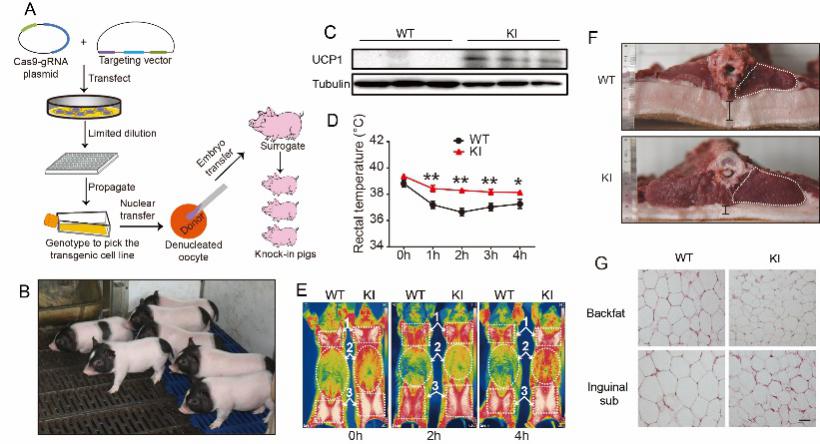
利用CRISPR/Cas9基因编辑技术在巴马猪中实现UCP1基因的定点插入,获得的UCP1 knockin猪表现为抗寒能力提高和瘦肉率增加,为低脂抗寒新品系培育提供新的素材。
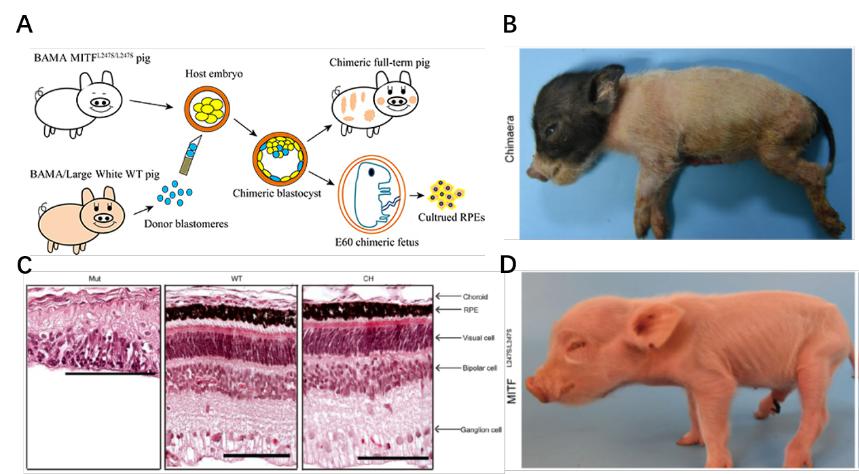
利用囊胚补偿技术首次在猪上实现眼睛的异体再造,为人类异种器官再生提供技术支持和动物模型,为眼睛的发育提供理论基础以及为眼睛疾病的治疗提供新的治疗方案。
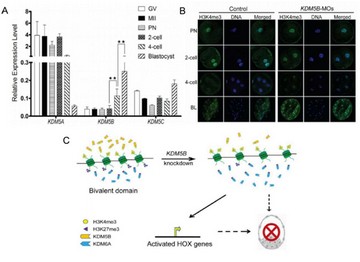
揭示组蛋白去甲基化酶KDM5B能够通过影响H3K4me3-H3K27me3的共价平衡改善HOX家族基因的激活,最终影响猪早期胚胎的发育。
| 研究内容和目标:
改造猪的基因组,培育猪的新品种,挖掘猪的新基因,构建生物医学研究的猪模型。 代表性发表论文: - Wang X§, Cao C§, Zhang Y, Jia Q, Zheng Q, Zhang H, Song R, Li Y, Luo A, Hong Q, Qin G, Yao J, Zhang N, Wang Y, Zhou Q, Zhao JG*. A harlequin ichthyosis pig model with a novel ABCA12 mutation can be rescued by acitretin treatment. J Mol Cell Biol, 2019, 11 (12), 1029-1041. (Cover story)
- Zhao JG§, Lai L, Ji W, Zhou Q*. Genome editing in large animals: Current status and future prospects. Natl Sci Rev, 2019, 6: 402-420. (Invited review)
- Cao C§, Zhang Y§, Jia Q, Wang X, Zheng Q, Zhang H, Song R, Li Y, Luo A, Hong Q, Qin G, Yao J, Zhang N, Wang Y, Wang H,Zhou Q, Zhao JG*. An exonic splicing enhancer mutation in DUOX2 causes aberrant alternative splicing and severe congenital hypothyroidism in Bama pigs. Dis Model Mech, 2019, 12: dmm036616.
- Zhang Y§, Hong Q, Cao C, Yang L, Li Y, Hai T, Zhang H, Zhou Q, Sui R*, Zhao J*. A novel porcine model reproduces human oculocutaneous albinism type II. Cell discovery, 2019, 5: 48.
- Zhang H§, Huang J§, Li Z§, Qin G, Zhang N, Hai T, Hong Q, Zheng Q, Zhang Y, Song R, Yao J, Cao C, Zhao JG*, Zhou Q*. Rescuing ocular development in an anophthalmic pig by blastocyst complementation. EMBO Mol Med. 2018;10(12). pii: e8861.
- Wang Y§*, Huang JJ, Zhao JG*. Gene engineering in swine for agriculture. J Integr Agr. 2017; 16(12): 2792-2804.
- Hai T§, Guo W§, Yao J§, Cao C, Luo A, Qi M, Wang X, Wang X. Huang J, Zhang Y, Wang D, Shang H, Hong Q, Zhang R, Jia Q, Zheng Q, Qin G, Li Y, Zhang T, Jin W, Chen ZY, Wang H, Zhou Q, Meng A, Wei H*, Yang S*, Zhao JG*. Creation of miniature pig model of human Waardenburg syndrome type 2A by ENU mutagenesis. Human genetics. 2017; 136:1463-1475.
- Zheng Q§, Lin J§, Huang J§, Zhang H, Zhang R, Zhang X, Cao C, Hambly C, Qin G, Yao J, Song R, Jia Q, Wang X, Li Y, Zhang N, Piao Z, Ye R, Speakman JR, Wang H, Zhou Q, Wang Y*, Jin W*, Zhao JG*. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decrease fat deposition and improve thermogenic capacity. Proc Natl Acad Sci U S A. 2017; 114(45): E9474-E9482.
- Zhang Y§, Xue Y§, Cao C§,Huang J, Hong Q, Hai T, Jia Q, Wang X, Qin G, Yao J, Wang X, Zheng Q, Zhang R, Li Y, Luo A, Zhang N, Shi G, Wang Y, Ying H, Liu Z, Wang H, Meng A, Zhou Q, Wei H*, Liu F*, Zhao JG*. Thyroid hormone regulates hematopoiesis via the TR-KLF9 axis. Blood. 2017; 130(20):2161-2170.
- Huang J§, Wang Y, Zhao J*. CRISPR editing in biological and biomedical investigation. J Cell Physiol. 2018 May;233(5):3875-3891
- Hai T§, Cao C§, Shang H§, Guo W§, Mu Y§, Yang S, Zhang Y, Zheng Q, Zhang T, Wang X, Liu Y, Kong Q, Li K, Wang D, Qi M, Hong Q, Zhang R, Wang X, Jia Q, Wang X, Qin G, Li Y, Luo A, Jin W, Yao J, Huang J, Zhang H, Li M, Xie X, Zheng X, Guo K, Wang Q, Zhang S, Li L, Xie F, Zhang Y, Weng X, Yin Z, Hu K, Cong Y, Zheng P, Zou H, Xin L, Xia J, Ruan J, Li H, Zhao W, Yuan J, Liu Z, Gu W, Li M, Wang Y, Wang H*, Yang S*, Liu Z*, Wei H*, Zhao JG*, Zhou Q*, Meng A*. Pilot study of large-scale production of mutant pigs by ENU mutagenesis. Elife. 2017 Jun 22; 6. pii: e26248.
- Lin J§, Cao C§, Tao C§, Ye R, Dong M, Zheng Q, Wang C, Jiang X, Qin G, Yan C, Li K, Speakman JR, Wang Y*, Jin W*, Zhao JG*. Cold adaptation in pigs depends on UCP3 in beige adipocytes. J Mol Cell Biol. 2017 Oct 1; 9(5):364-375.
- Jia Q§, Cao C§, Tang H, Zhang Y, Zheng Q, Wang X, Zhang R, Wang X, Luo A, Wei H, Meng A, Zhou Q, Wang H*, Zhao JG*. A 2-bp insertion (c.67_68insCC) in MC1R causes recessive white coat color in Bama miniature pigs. J Genet Genomics. 2017 Apr 20; 44(4):215-217.
- Yao J§, Huang J, Zhao JG*. Genome editing revolutionize the creation of genetically modified pigs for modeling human diseases. Hum Genet. 2016 Sep;135(9):1093-105.
- Wang X§, Cao C§, Huang J§, Yao J, Hai T, Zheng Q, Wang X, Zhang H, Qin G, Cheng J, Wang Y, Yuan Z, Zhou Q, Wang H*, Zhao JG*. One-step generation of triple gene targeted pigs using CRISPR/Cas9 system. Sci Rep. 2016; 6:20620.
- Huang J§, Zhang H, Yao J, Qin G, Wang F, Wang X, Luo A, Zheng Q, Cao C, Zhao JG*. BIX-01294 increases pig cloning efficiency by improving epigenetic reprogramming of somatic cell nuclei. Reproduction. 2016 Jan; 151(1):39-49.
- Zhao JG§. , Xu WJ, Ross JW., Walters EM., Butler SP., Whyte JJ., Kelso L., Fatemi M, Vanderslice NC., Giroux K., Spate LD., Samuel MS., Murphy CN., Wells KD., Masiello NC., Prather RS*. & Velander WH*. Engineering protein processing of the mammary gland to produce abundant hemophilia B therapy in milk. Sci Rep. 2015 Sep 21; 5: 14176.
- Wang X§, Zhou J§, Cao C§, Huang J, Hai T, Wang Y, Zheng Q, Zhang H, Qin G, Miao X, Wang H, Cao S*, Zhou Q*, Zhao JG*. Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Sci Rep. 2015 Aug 21; 5: 13348.
- Zhao W§, Wang Y§, Liu S, Huang J, Zhai Z, He C, Ding J, Wang J, Wang H, Fan W, Zhao JG*, Meng H*.The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One. 2015, 10(2): e0117441.
- Huang J§, Zhang H, Wang X, Dobbs KB, Yao J, Qin G, Whitworth K, Walters EM, Prather RS*, Zhao JG*. Impairment of Preimplantation Porcine Embryo Development by Histone Demethylase KDM5B Knockdown Through Disturbance of Bivalent H3K4me3-H3K27me3 Modifications. Biol Reprod. 2015, 92(3):72. (*Co-corresponding Author)
- Yao J§, Huang JJ, HaiTang , Wang XL, Qin GS, Zhang HY, Wu R, Xi J. J, Yuan Z, Zhao JG*. Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs. Sci Rep. 2014, 4:6926. (*Corresponding Author)
- Wang YP§, Qi ST, Wei Y, Ge ZJ, Chen L, Hou Y, Ouyang YC, Schatten H, Zhao JG*, Sun QY*.Knockdown of UCHL5IP causes abnormalities in γ-tubulin localisation, spindle organisation and chromosome alignment in mouse oocyte meiotic maturation. Reprod Fertil Dev. 2013; 25(3):495-502. (*Co-corresponding Author)
- Whyte JJ§,Zhao JG§, Samuel M, Laughlin HM, Prather RS*, Gene Targeting with Zinc Finger Nucleases to Produce Cloned eGFP Knockout Pigs. Mol Reprod Dev. 2011, 78(1):2.
- Zhao JG§, Whyte JJ, Prather RS*. The effect of epigenetic regulation on somatic cell nuclear transfer. Cell. Cell Tissue Res. 2010, 341(1):13-21.
- Zhao JG§, Hao Y, Ross JW, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS*. Histone Deacetylase Inhibitors Improve In Vitro and In Vivo Developmental Competence of Somatic cell Nuclear Transfer Porcine Embryos.Cloning and stem cells,2010,12(1):75-83.
- Ross JW§, Prather RS, Whyte JJ,Zhao JG*.Cloning and transgenics: progress and new approaches.Invited review by Centre for Agricultural Bioscience International,2009, 4, No. 38.
- Zhao J§, Ross JW, Hao Y, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS*. Significant Improvement in Cloning Efficiency of an Inbred Miniature Pig by Histone Deacetylase Inhibitor Treatment after Somatic Cell Nuclear Transfer. Biol Reprod. 2009, 81: 525-530.

|

